1.SNVs (single nucleotide variants)
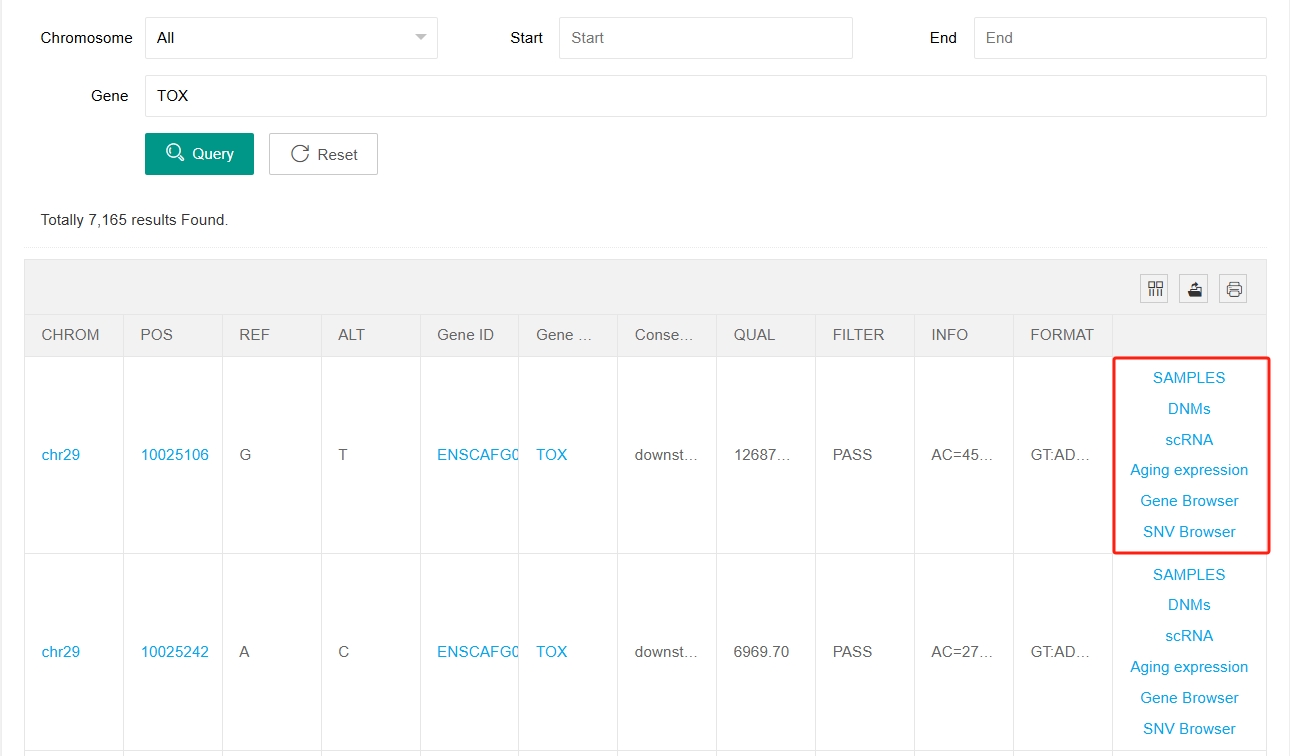
The posted Dog10K SNV VCF file has been integrated into a visualised table. Users can enter the chromosome number, start and end site to search for variation information at the corresponding site.
The posted Dog10K SNV (single nucleotide variant) VCF file has been integrated into a visualised table.
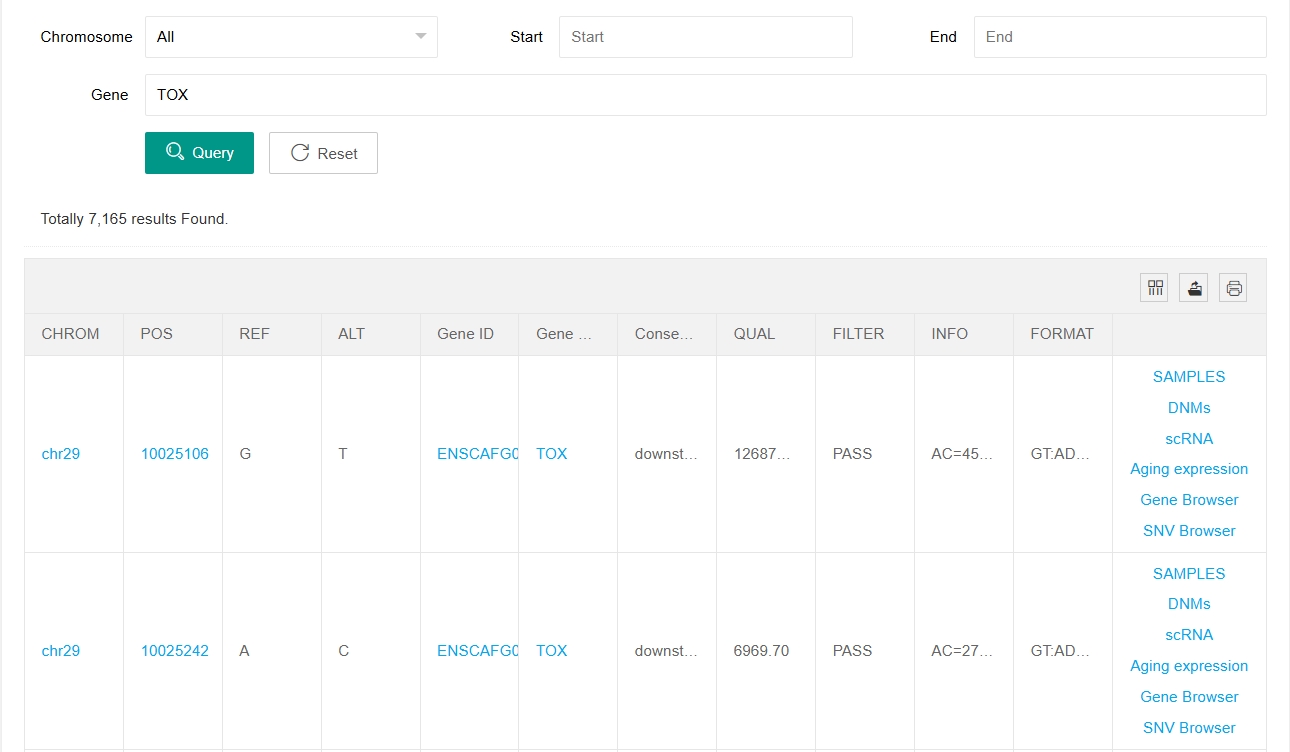
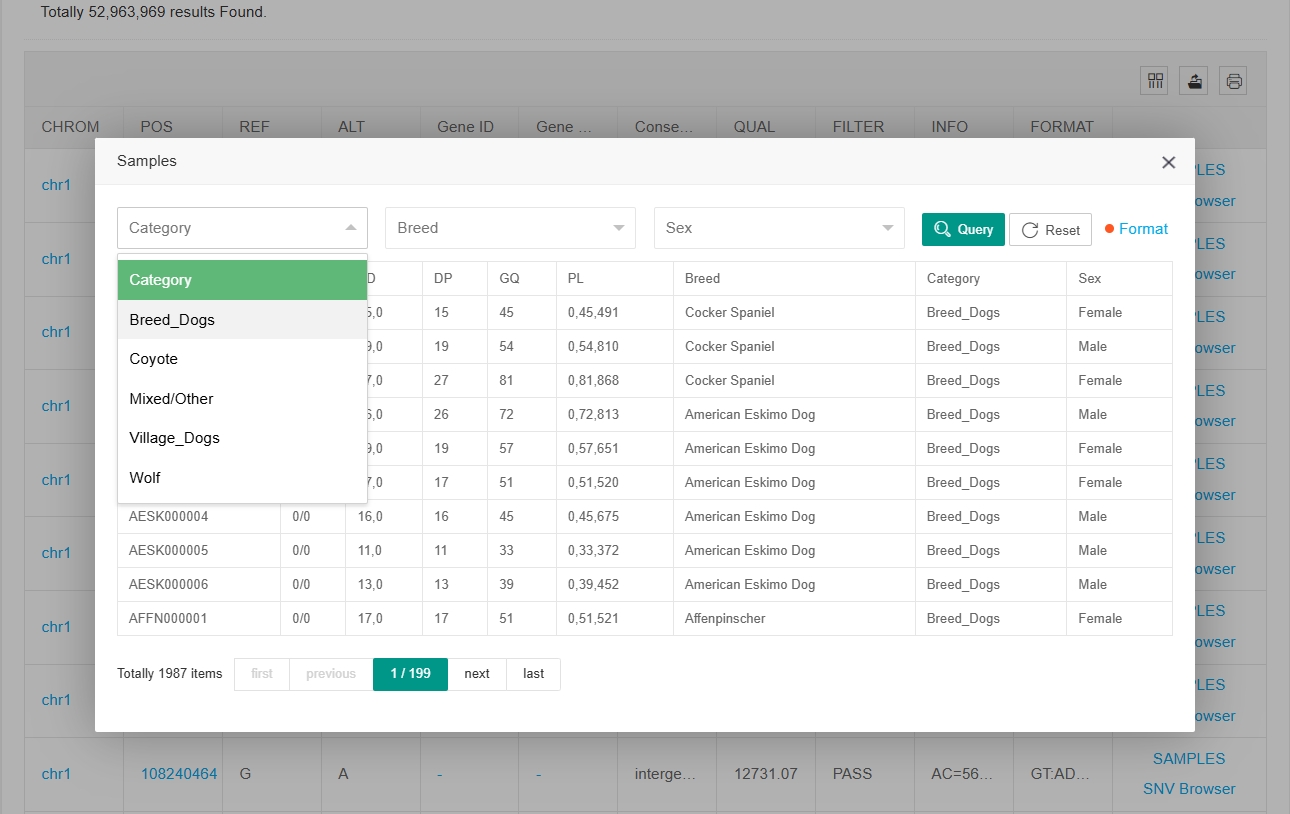
By clicking on “SNV Browser” or “Gene Browser” in the last column, users can jump to the Genome Browser web page for that site or gene. Click on "SAMPLES" in the last column of each site and a new window will open showing variation and sample information for each individual at that site. In this window you can filter for the breeds and sexes you are interested in.
2.DNMs (de-novo mutations)
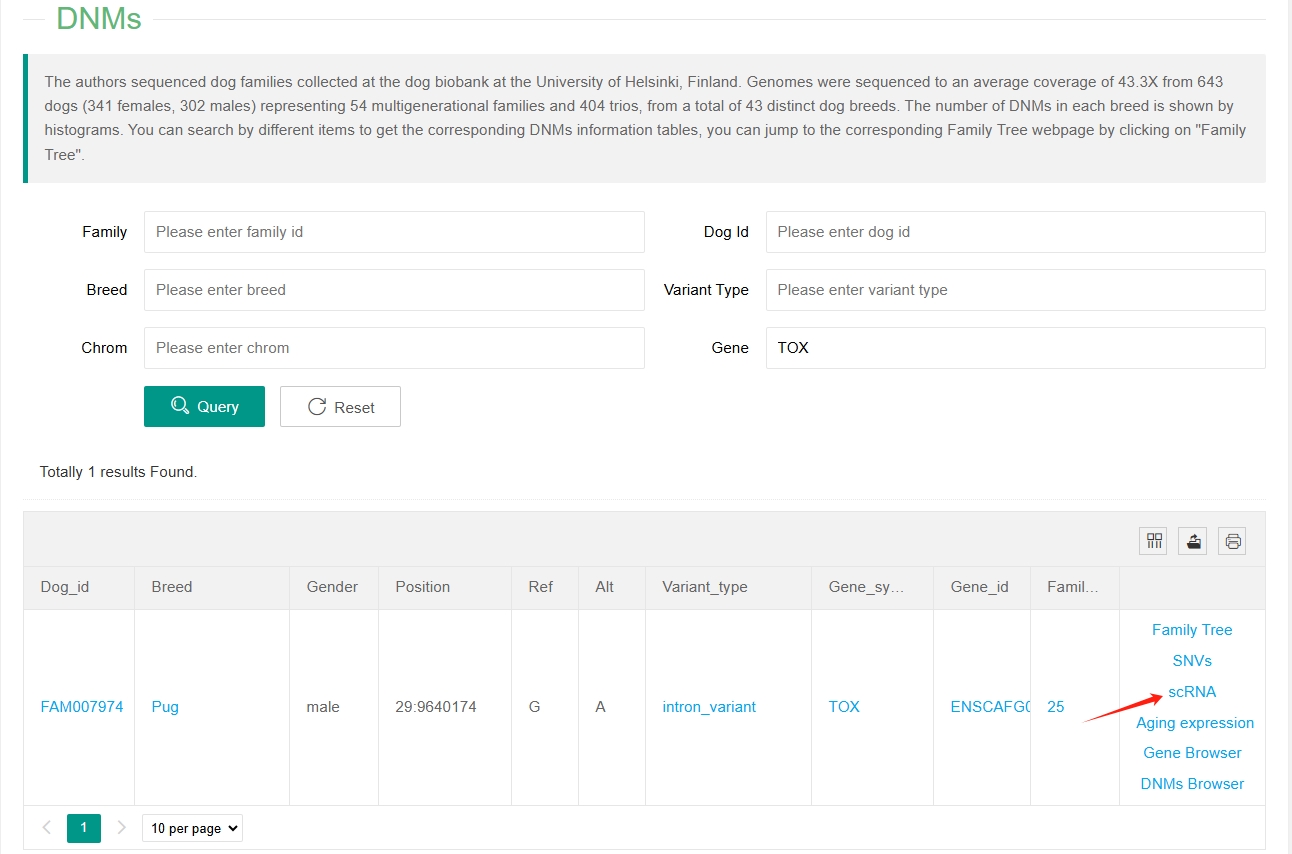
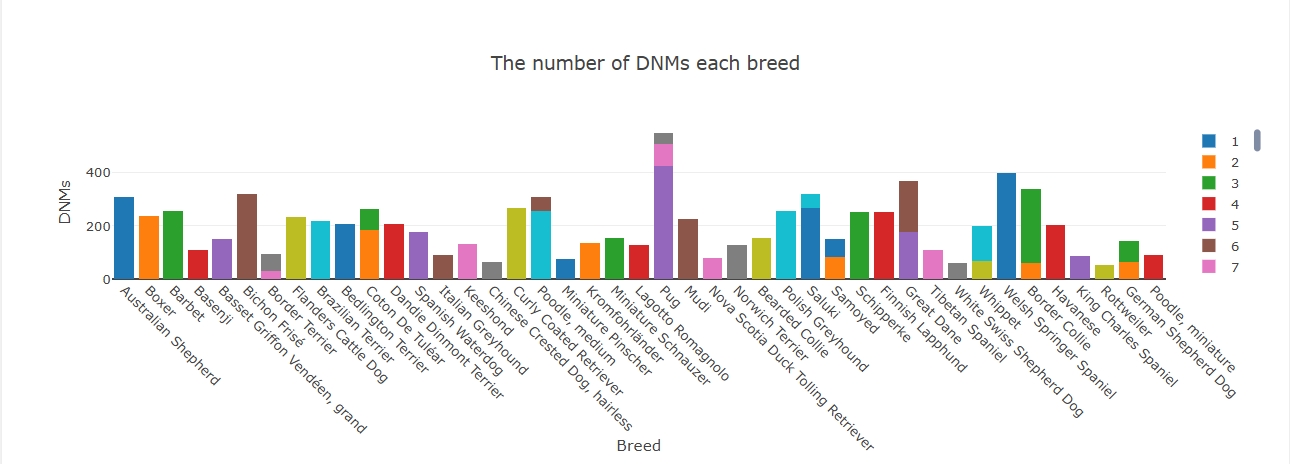
The number of DNMs (de-novo mutations) in each breed is shown by histograms, different colours mean different families, one or more families may be included in a breed.
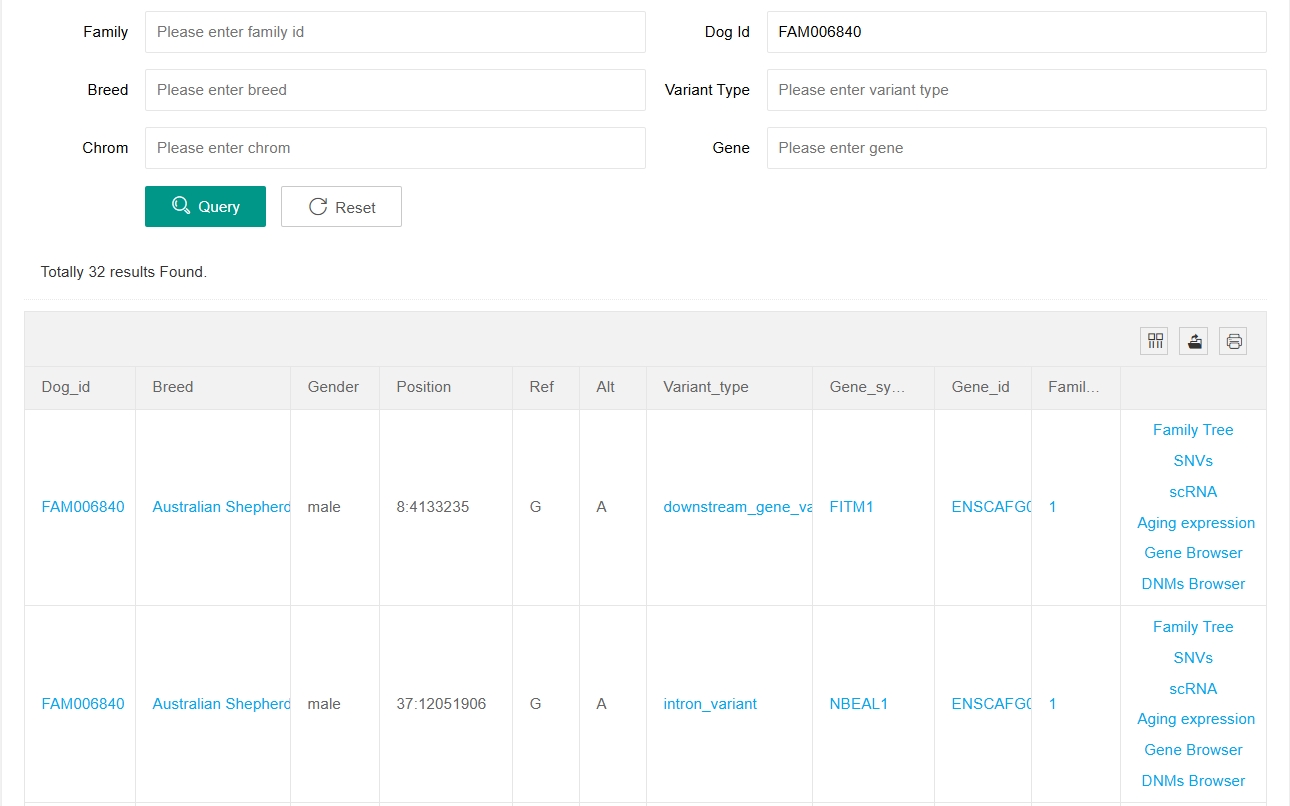
Below the histograms is the mutation information table for each individual and the search function. Users can enter Family ID, Dog ID, Breed, Variant Type, Chromsome Number and Gene to search for relevant mutation information. Users can also click on a family in the upper histogram to search for mutation information for that family. Clicking on items in the table can also search for the DNM information of the corresponding items.
In addition, in order to display the relationship among individuals intuitively, we also built the family tree for each Family. Users can click on the "Family Tree" in the last column of the table and a new window will open with the corresponding family tree.
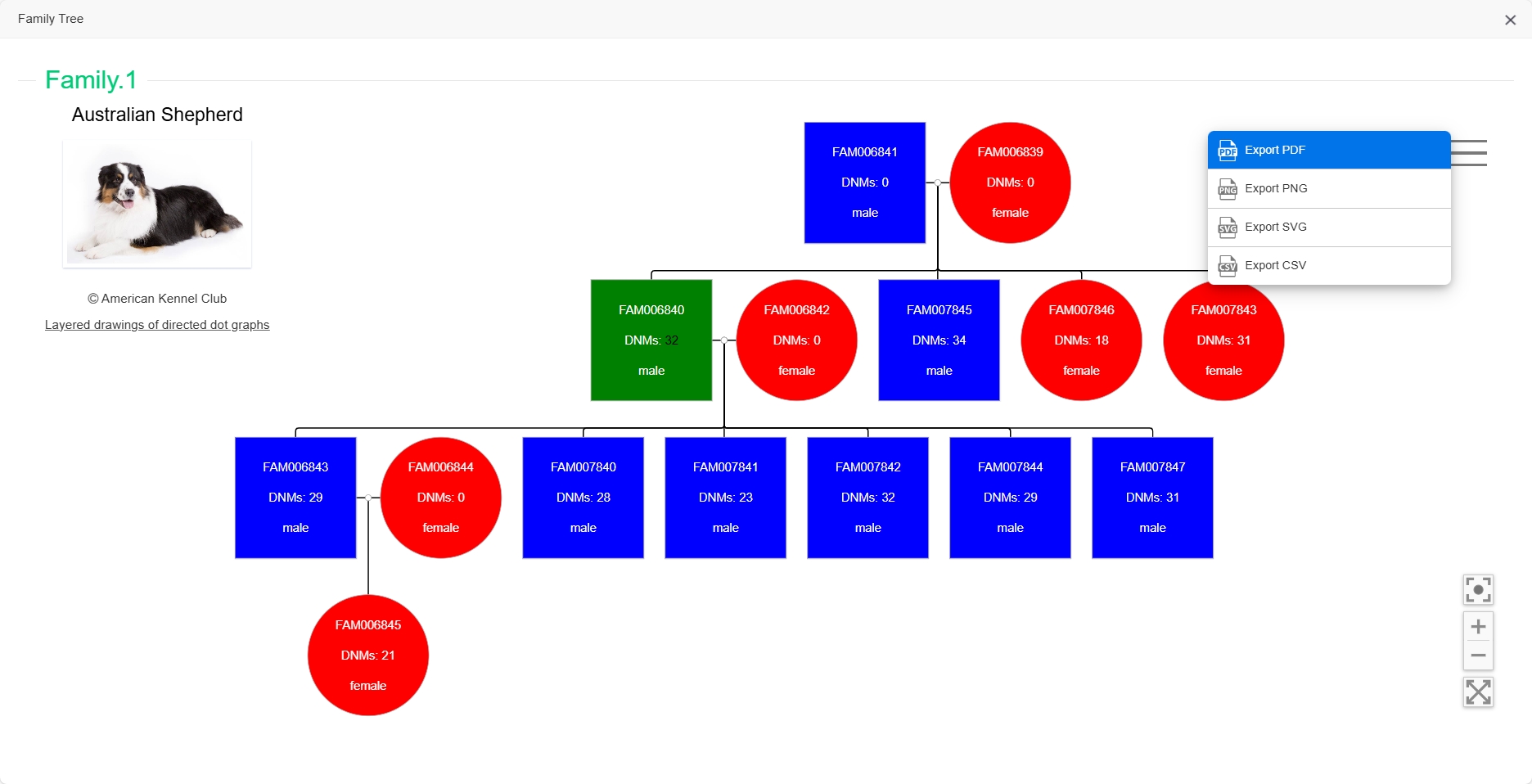
The Family Tree webpage included the breed photo and family tree of this family, the text in each box shows the individual's sample name, number of DNMs, and sex. Users can also export the family trees to various file types.
3.scRNA (single-cell RNA sequencing)
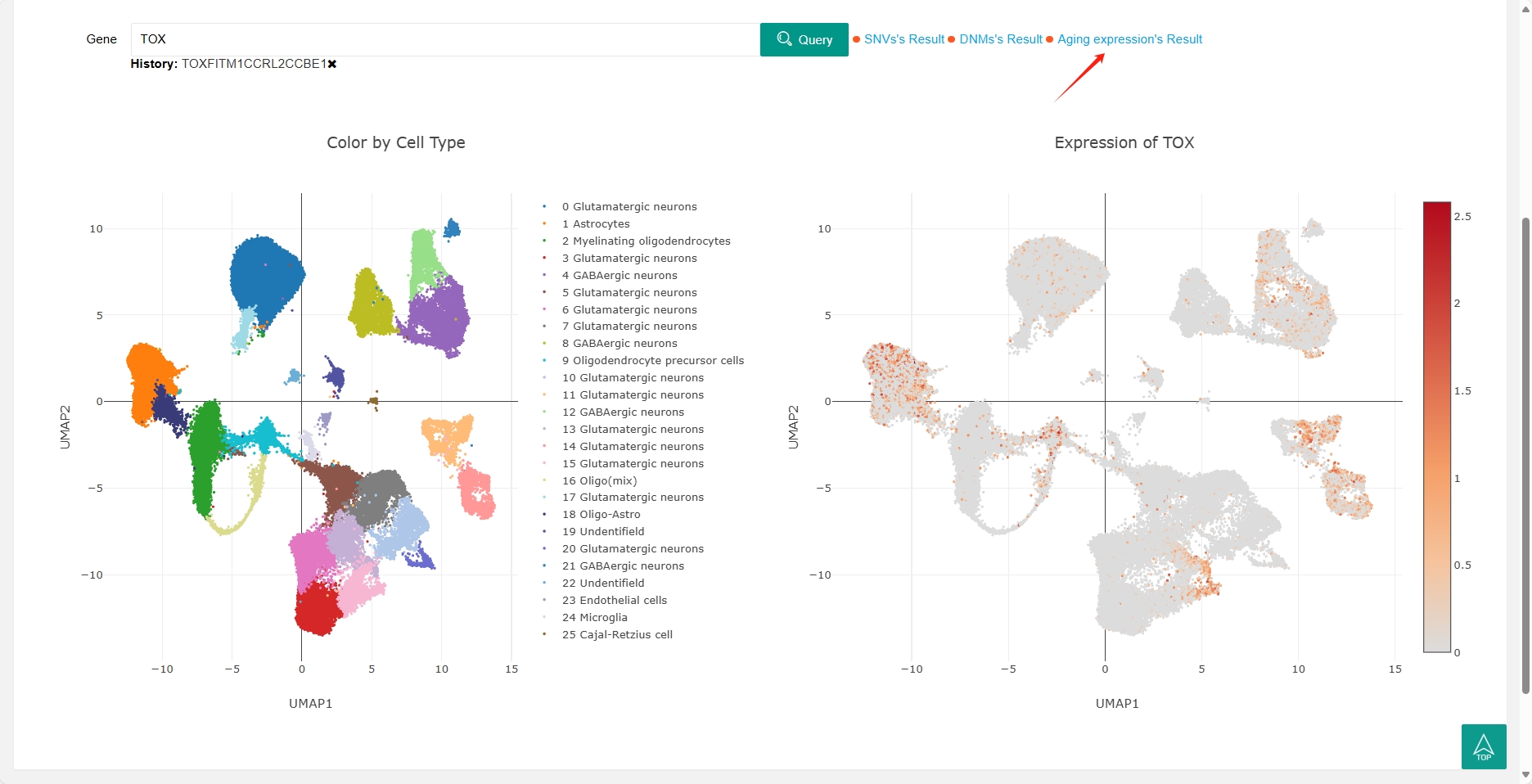
The scRNA (single-cell RNA sequencing) section now includes two single cell studies in dogs, users can search for genes to view the gene's expression profile in each cluster (right). The cell types corresponding to each cluster are also shown (left).
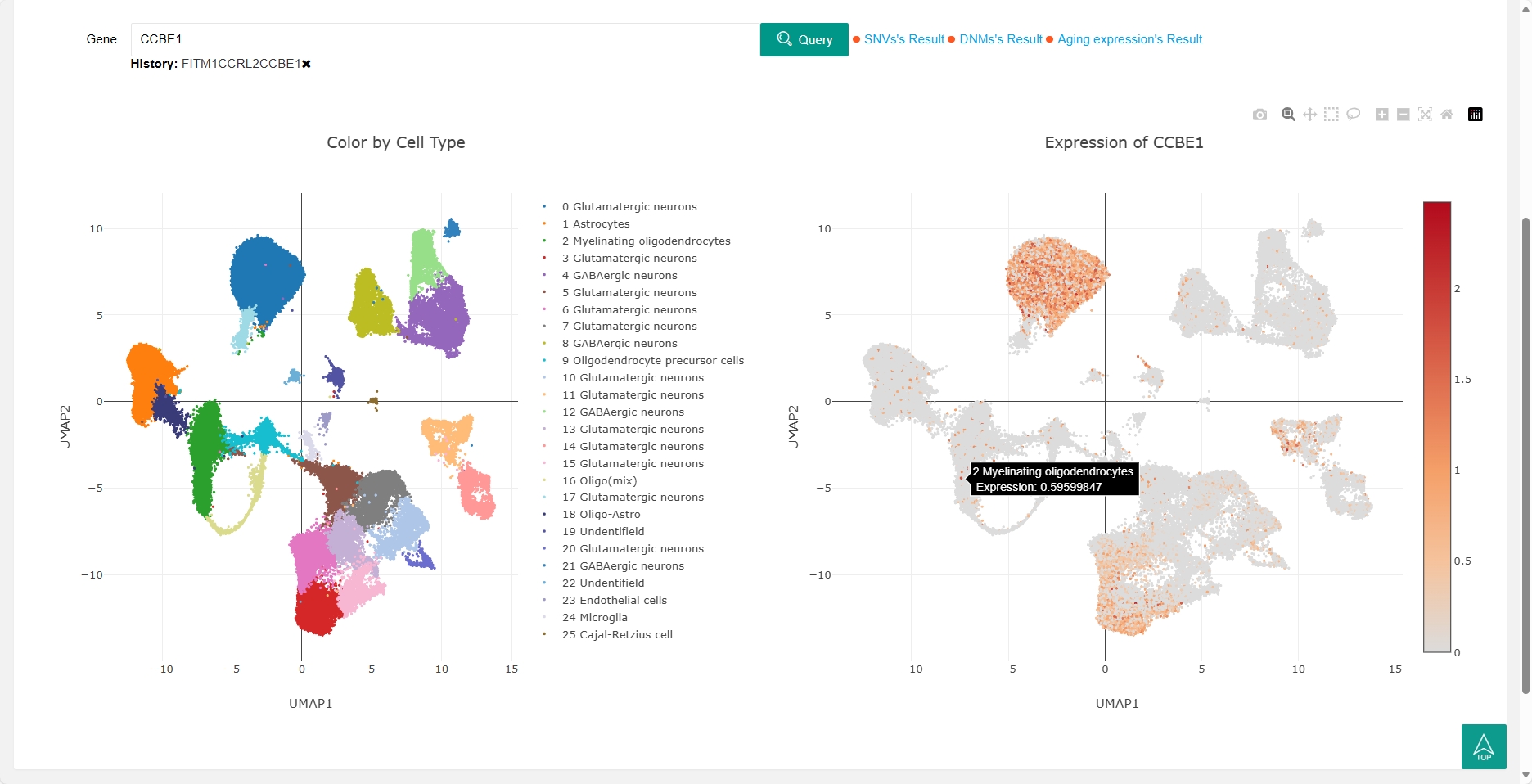
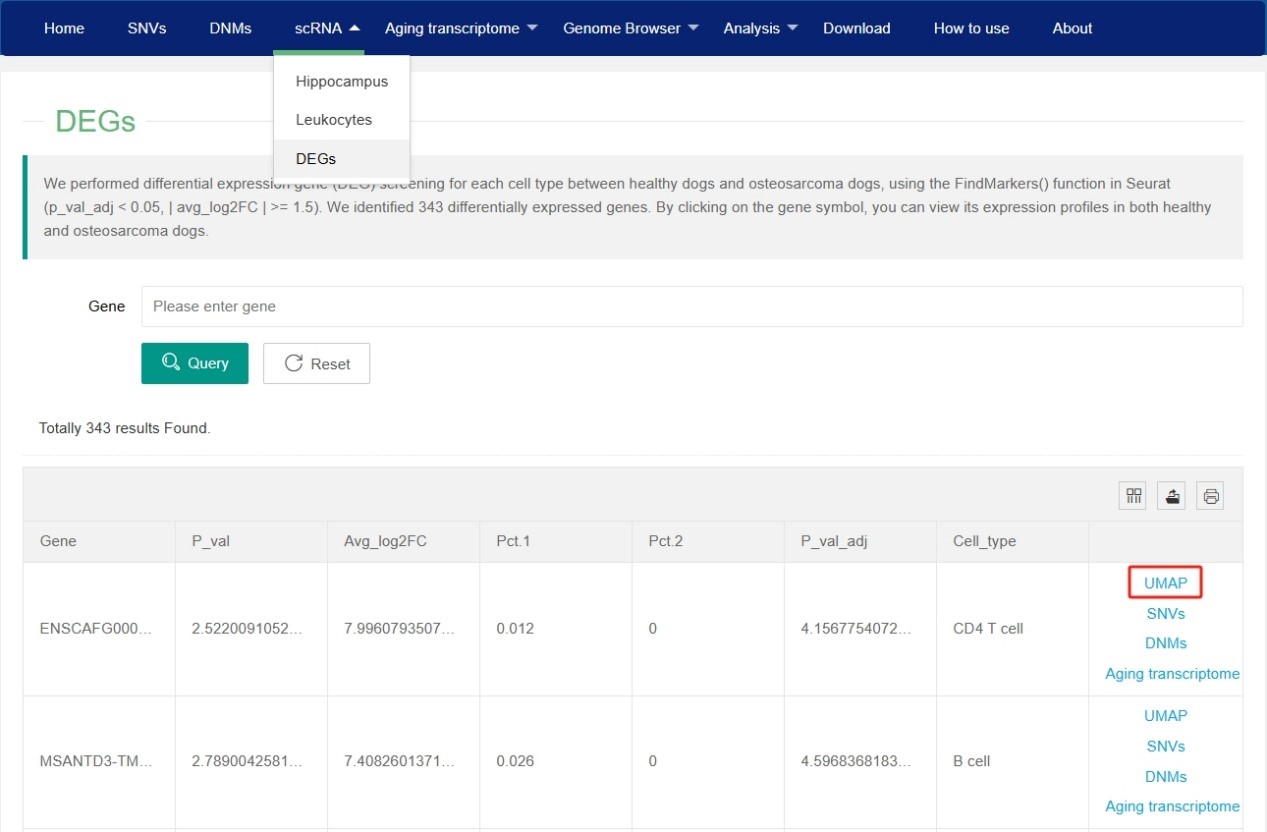
In the ‘DEGs’ of scRNA section, users can view the differential expression genes (DEGs) for each cell type between healthy dogs and osteosarcoma dogs. By clicking on ‘UMAP’ in the last column of the DEGs table, users can view the expression profile of this gene in both healthy dogs and osteosarcoma dogs.
4.Aging transcriptome
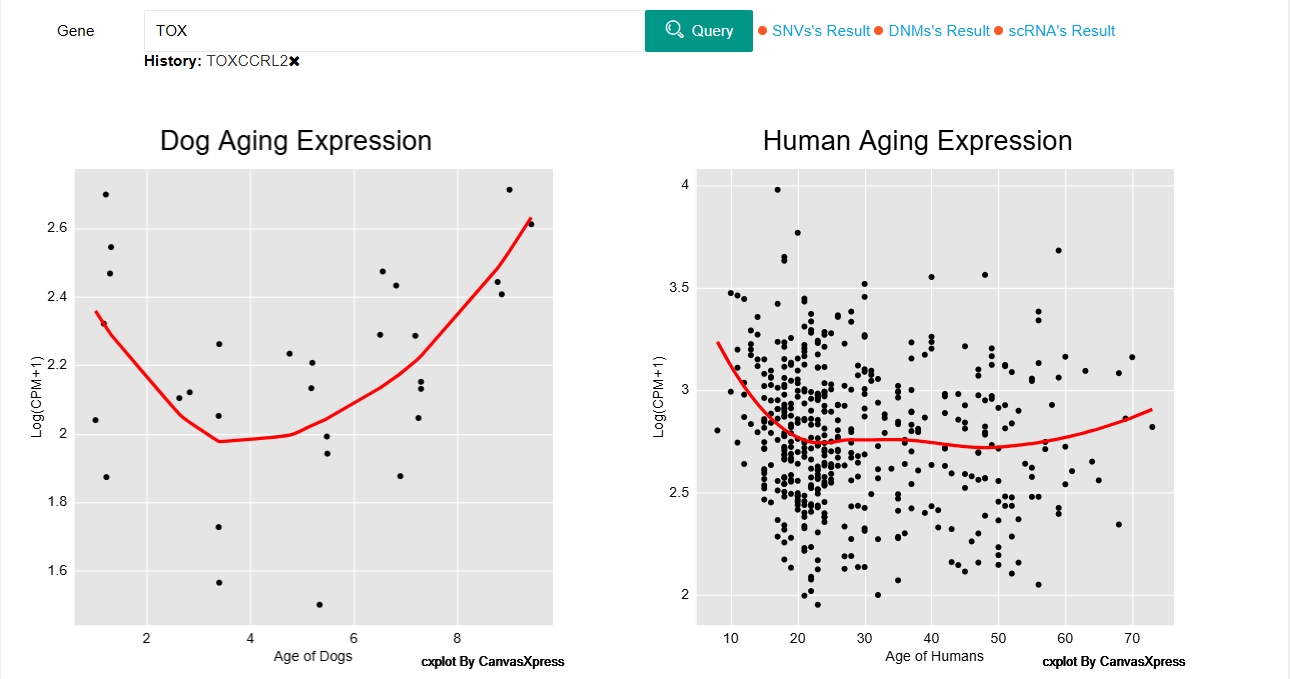
We conducted scatter plots using log-transformed counts per million (CPM) for humans and dogs of serial ages, and applied the loess method to smooth the fitted curves. Users can search for the gene of interest and view its expression profile in dogs and humans of different ages.
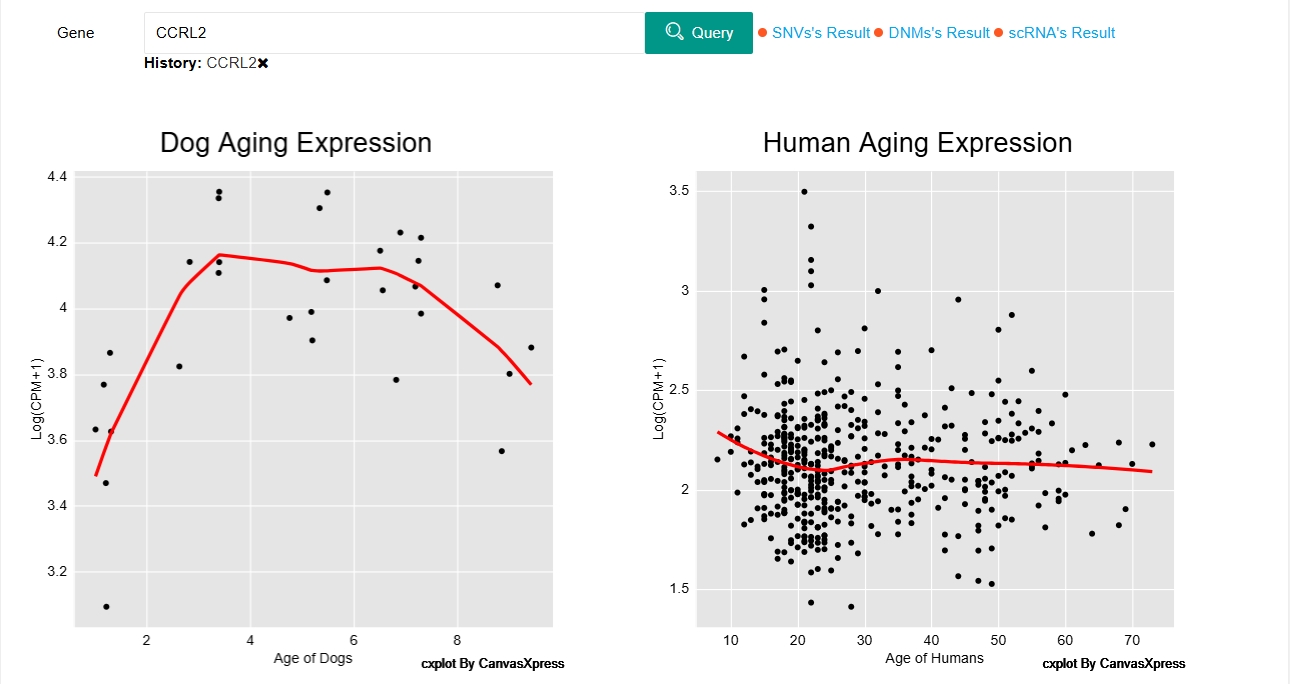
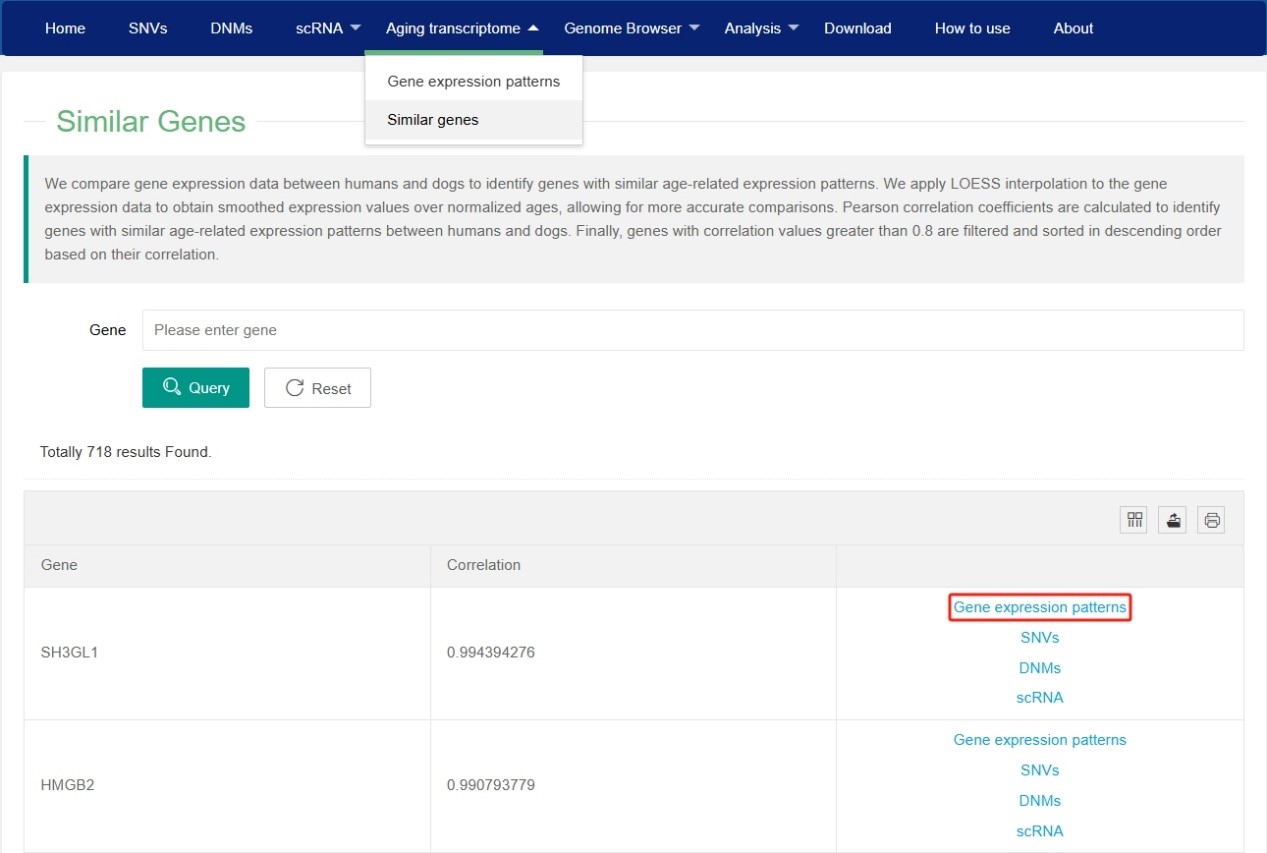
In the ‘similar genes’ of Aging transcriptome section, users can view the genes with similar age-related expression patterns between humans and dogs. By clicking on ‘Gene expression patterns’ in the last column of the similar genes table, users can view the expression profile of this gene in both dogs and humans at different ages.
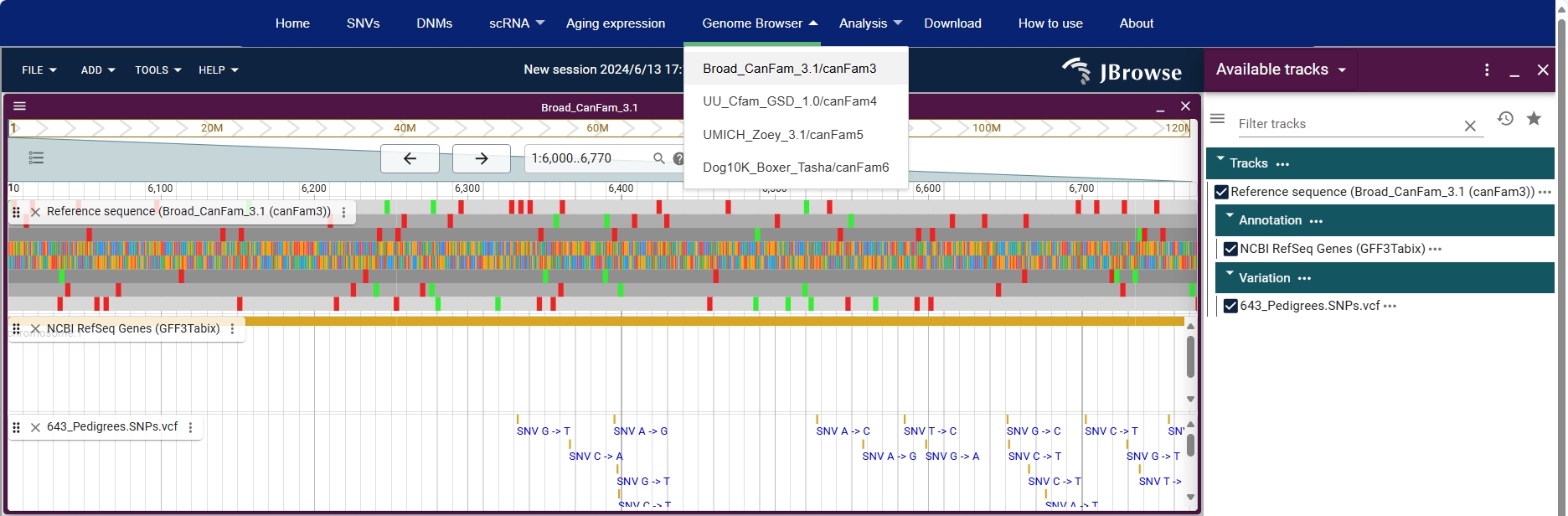
5.Genome Browser
Genome Browser section provides visualisation of the four releases of the canine genome and annotation. In the Broad_CanFam_3.1/canfam3 release, you can browse the vcf file for DNMs studies (643_Pedigrees.SNPs.vcf). In the UU_Cfam_GSD_1.0 /canFam4 release, you can browse the vcf file for Dog10K (DOG10K.SNPs.vcf).
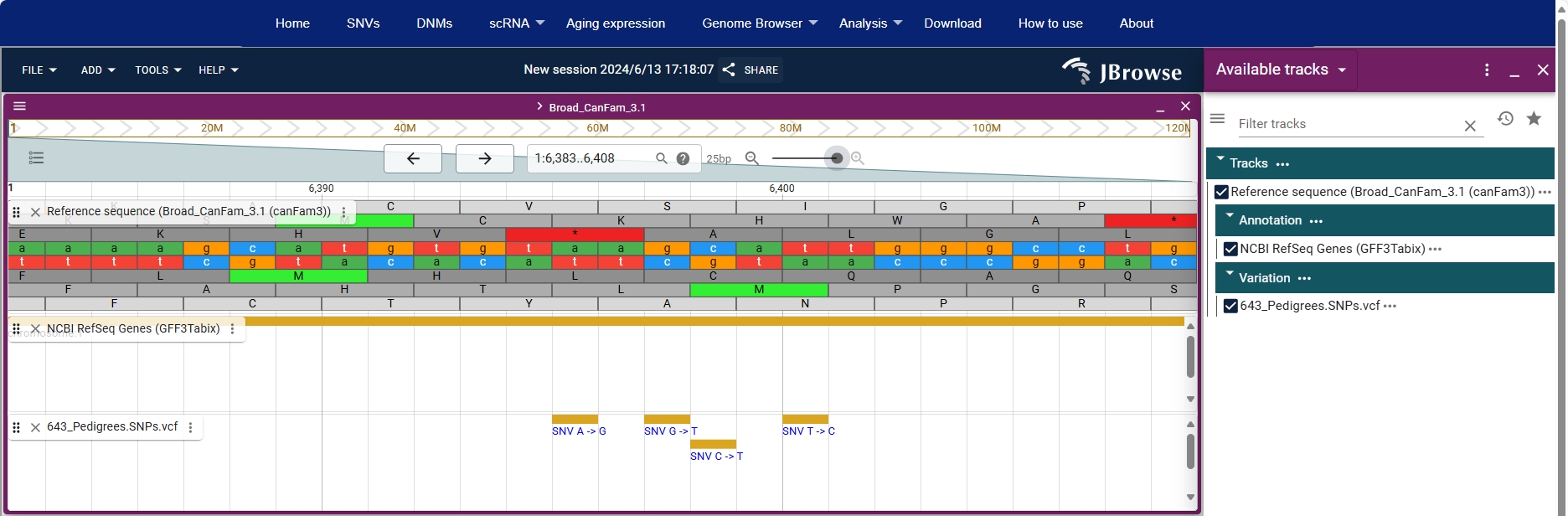
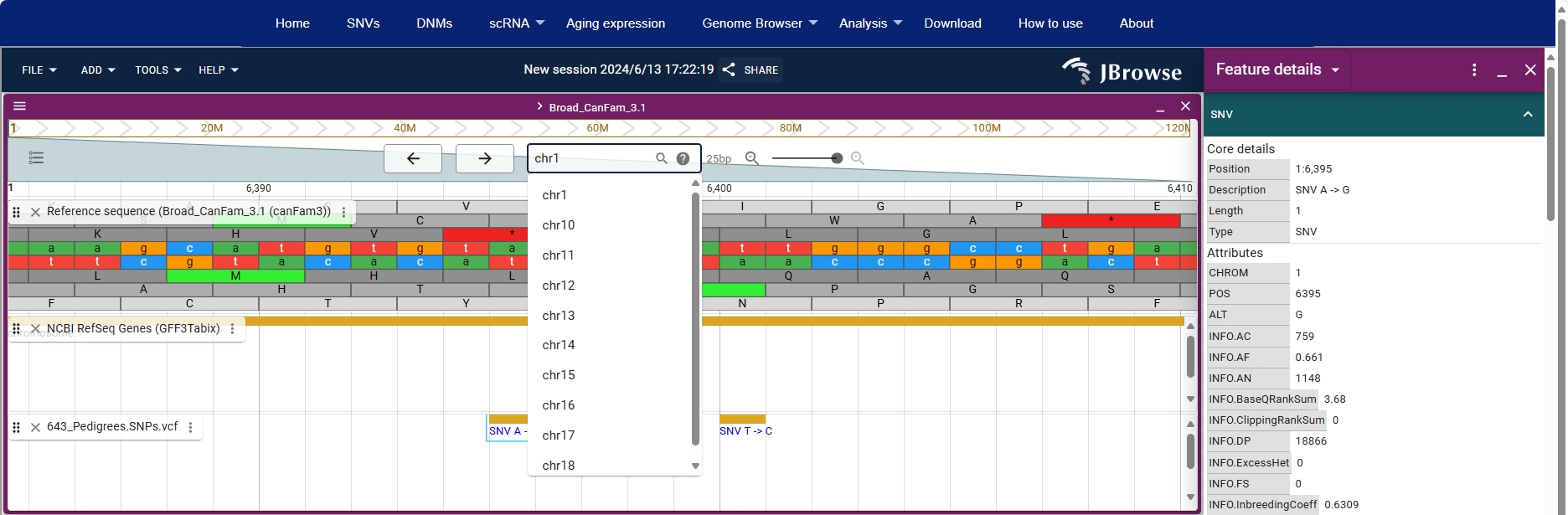
Users can zoom in to see the base sequence of the genome.
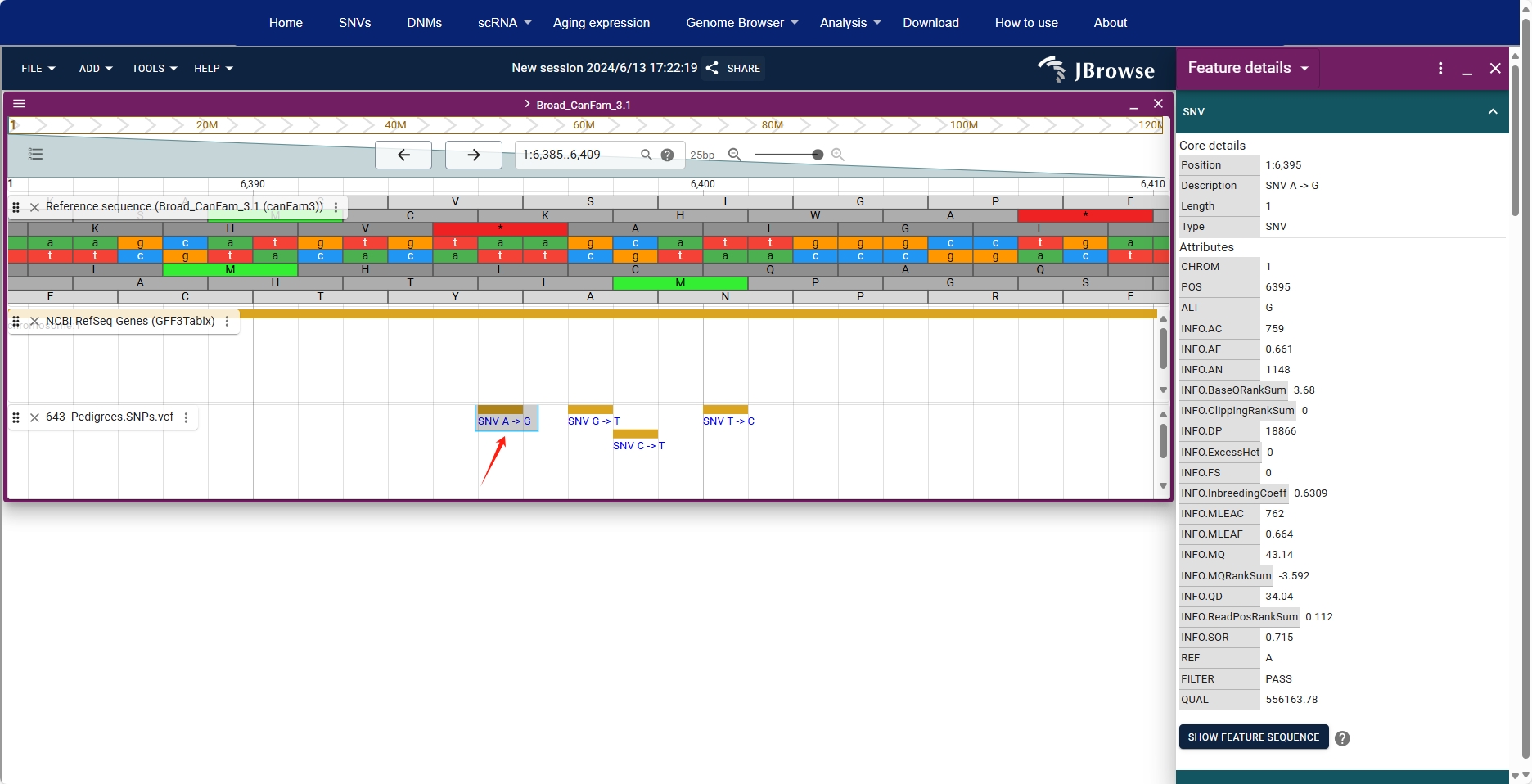
By clicking on a site in the VCF box, users can view detailed information about that site on the right.
Users can search for chromosomes (eg. chr1, 1, NC_006583.4), locations (eg. 1:5,361..7,304), genes (eg. Tox), etc. in the search box to browse the relevant information.
6.Analysis
Liftover
There are currently several versions of the dog genome. Researchers studying the human or mouse genome may also wish to view the variant information at the corresponding loci in dogs. We therefore provide a tool to convert positions between the human, dog and mouse genomes. This function is copied from UCSC's Liftover (https://genome.ucsc.edu/cgi-bin/hgLiftOver). Users can choose the Original Genome (Assembly) and the New Genome (Assembly), then enter text in BED format or submit a file in BED format (https://grch37.ensembl.org/info/website/upload/bed.html) for positional information conversion. The BED format consists of at least three columns: chromosome number, start position and end position (e.g. "chr1 10000 10088").
Selscan
Selscan is an efficient tool for Haplotype-based scan to detect natural selection, which is useful to identify recent or ongoing positive selection in genomes.
Users can first select the analysis method they wish to use in the ‘Method’ section (XP-nsl, XP-EHH, iHS, iHH12 and nSL).
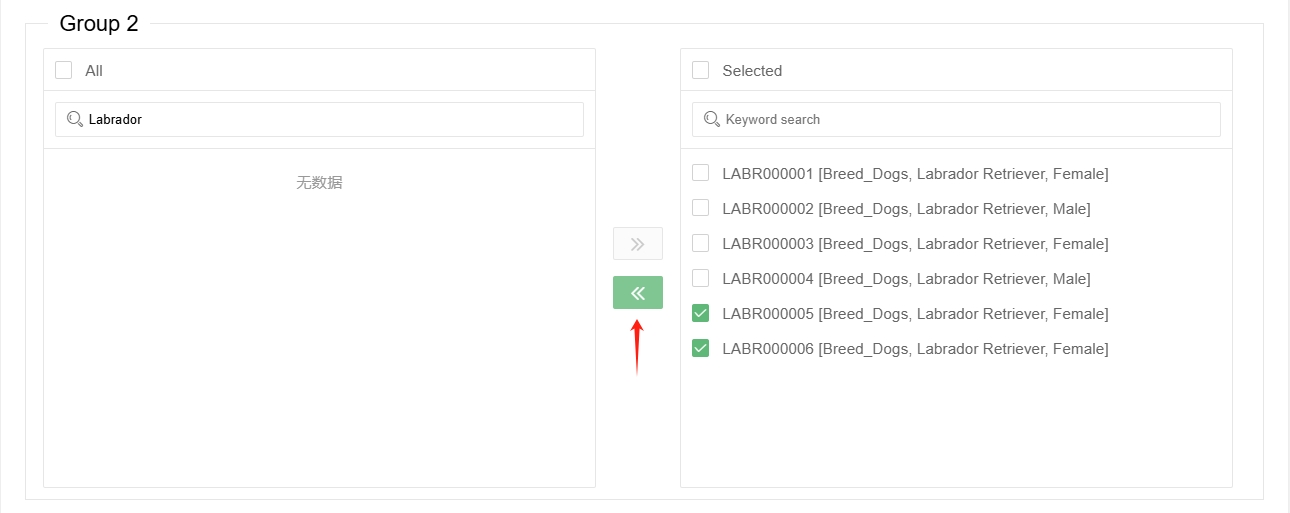
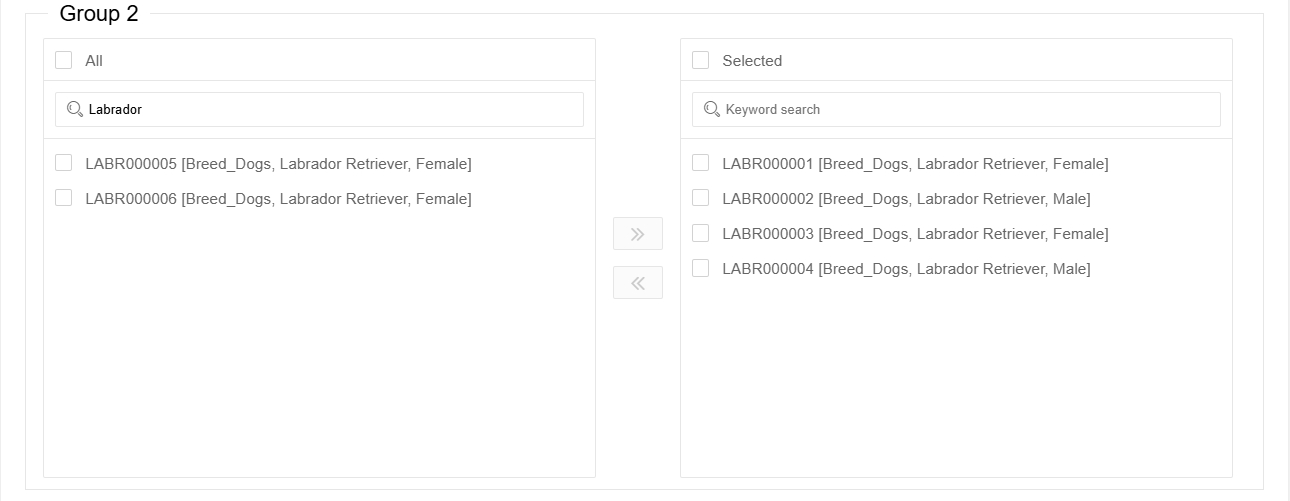
Then select the samples to be analysed in the 'Group' box below, you can use keyword search to filter samples of a particular category (note that the search is case-sensitive).
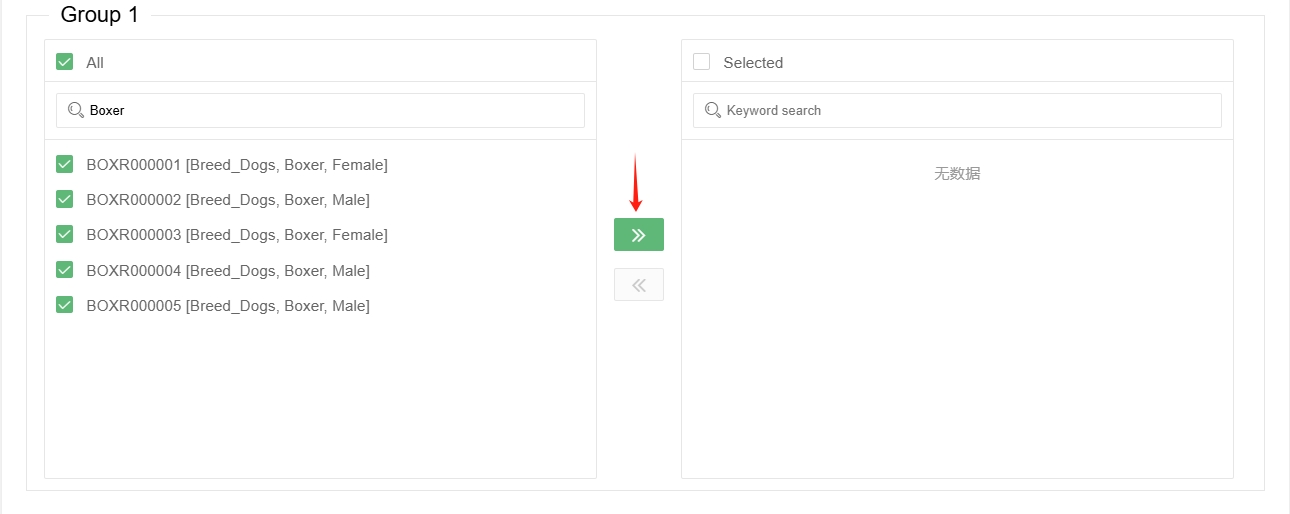
Click the ‘Go right’ button in the middle and add them to the ‘Selected’ box on the right.

Finally, enter the e-mail address and click 'Submit'. The Selscan selection analysis of the relevant group will be run on our server. Due to the long running time (several hours to several days) the final result will be sent to you by e-mail.
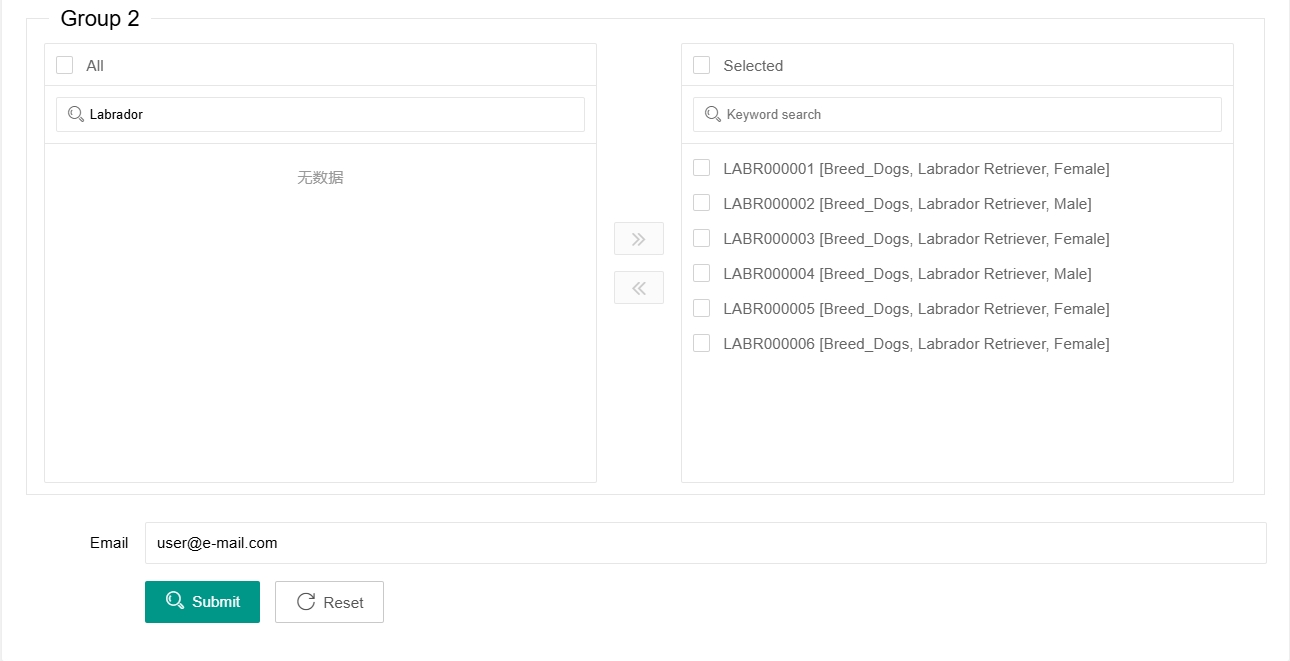
If the samples are in the 'Selected' box on the right, they have already been selected for analysis. It is not necessary to select them again in the right box. By selecting samples in the right box and clicking the 'Go left' button in the middle, you can remove these samples from the selected ones.
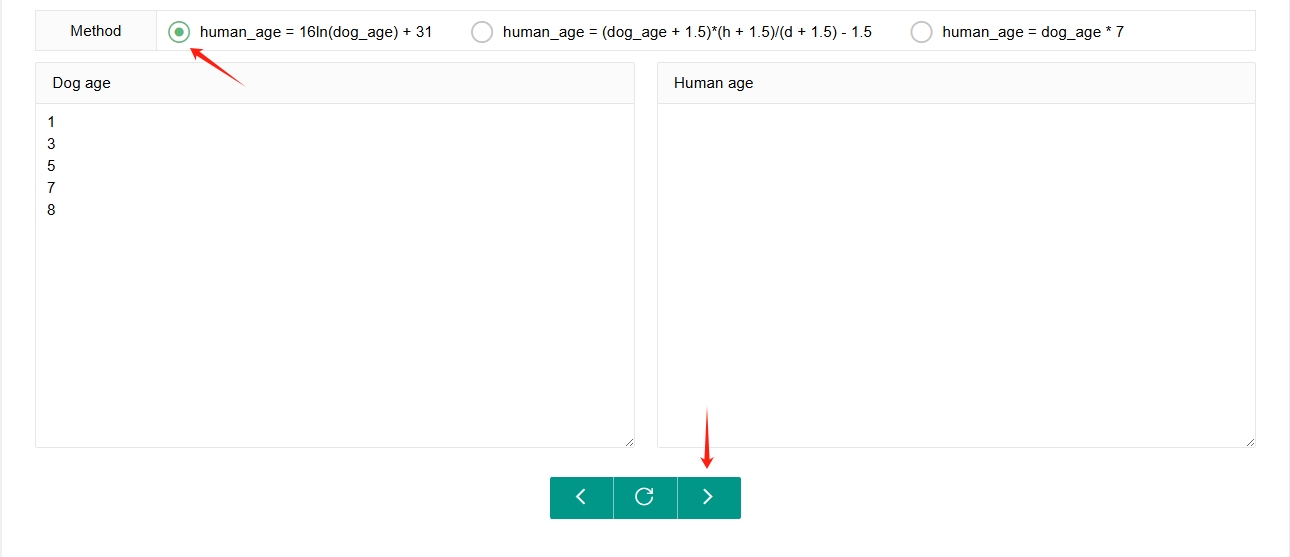
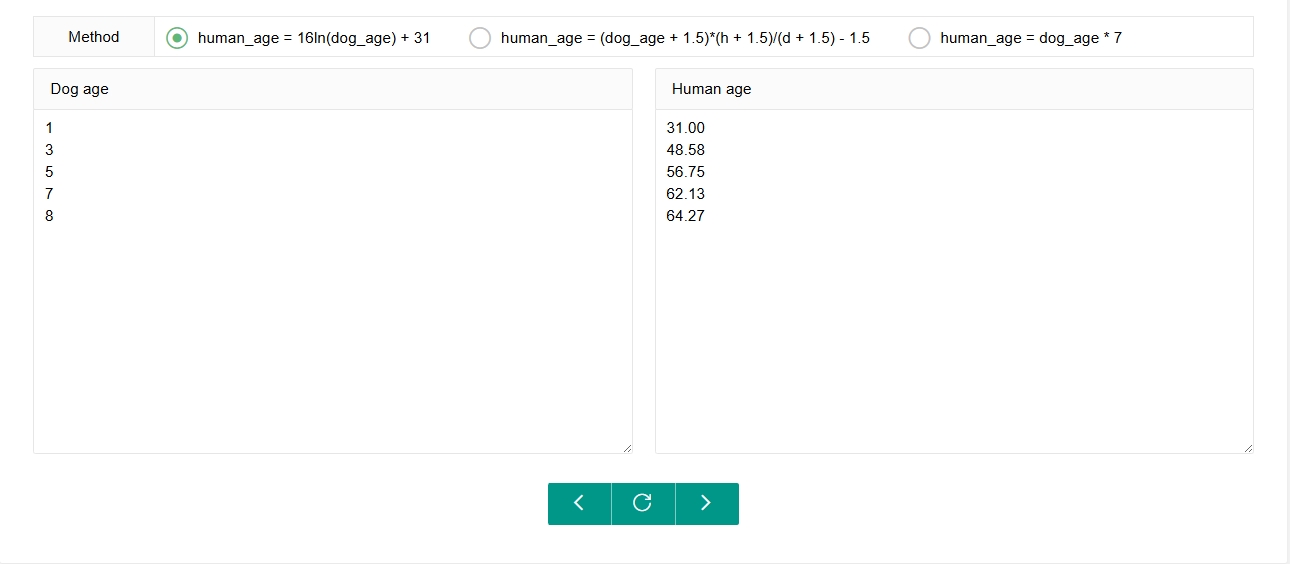
AgeConversion
We provide a small tool to make it easier for users to perform single or batch age conversions between dogs and humans. Users can select the age conversion formula they wish, then enter the age to be converted in either the ‘Dog age’ or ‘Human age’ box. By clicking on the arrow corresponding to the desired conversion direction, the converted result will display in the other box.
7.Download
The Download section provides the download of the data presented in the database. All these data have been published, and they were colleted from various articles and databases.
Variant files and associated annotations from Dog10K consortium are available at https://kiddlabshare.med.umich.edu/dog10K/ and at https://zenodo.org/record/8084059. Raw sequence data of DNMs by Zhang et al. is available at GSA (https://ngdc.cncb.ac.cn/gsa/) under accessions CRA004356 CRA002653 CRA002915 and CRA001113; and NCBI (https://www.ncbi.nlm.nih.gov/) under the Bioproject PRJNA1079355. The sn-RNA sequencing data by Zhou et al. is available at GSA (https://ngdc.cncb.ac.cn/gsa/) under accessions PRJCA004294. The sc-RNA sequencing data by Ammons et al. is available at the NCBI Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE225599. The dog aging transcriptome data by Zeng et al. are available at GSA (https://ngdc.cncb.ac.cn/gsa/) under accessions PRJCA016985. The human aging transcriptome data we downloaded are available at the NCBI Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE94438, GSE112087, GSE123658 and GSE134080.
8.Other notes
Joint analysis of multiple omics is the future trend, and most results in our database are interconnected. For instance, in the SNVs section, clicking on "DNMs"" in the last column of the table will direct you to the DNM information for that gene in the DNMs results. Similarly, in the DNMs section, clicking on "snRNA" in the last column of the table will take you to the expression profile of that gene in the single-cell results. Furthermore, in the snRNA section, you can click on "Aging expression's Result" next to the search box to view the gene's expression profile in the aging expression results.